Data Model
This section provides a quick tour of much of the data that came out of HMP1, however it is not a complete inventory of all data sets and analysis products. For that, see the legacy HMP1 Data Browser, providing access to specific HMP1 datasets described in 2012 and 2017 landmark Nature papers.
Framework of Sequence data: Cohort type and Data type
One way of organizing the metagenomic sequence data generated by HMP1 is to split it by cohort type and data type.
Two primary cohort types:
- Center "Healthy Cohort": This is a single cohort of 300 healthy individuals, each sampled at 5 major body sites (oral, airways, skin, gut, vagina) at up to three timepoints. Each body site consisted of a number of body subsites, for a total of 15 to 18 samples per individual per timepoint. Read more about 16S & metagenomic sampling and sequencing efforts.
- Demonstration Project "Disease Cohorts": These 15 projects each have one or more cohorts aimed at studying specific health conditions. Each project developed sampling, processing, and 16S or whole metagenome shotgun sequencing approaches according to their condition of interest. These cohorts include both controls and affected individuals. Read more about Demonstration projects.
Three primary data types:
- Reference microbial genomes: Most of these are not derived from specific cohorts
- Whole metagenome shotgun (mWGS) sequence
- 16S metagenomic sequence
The resulting division can be roughly represented by the following table:
| Center "Healthy Cohort" | Demonstration Project "Disease Cohorts" | |
| Reference microbial genomes | >3000 strains NCBI BioProject 28331 |
Hundreds of strains NCBI BioProject 46305 |
| mWGS metagenomic sequence | Subset of the 300 subjects, multiple timepoints, 15+ bodysites NCBI BioProject 43017 |
5 projects, each with unique, sampling sites, conditions, etc. NCBI BioProject 46305 |
| 16S metagenomic sequence | 300 subjects, multiple timepoints, 15+ bodysites NCBI BioProject 48489 |
14 projects, each with unique, sampling sites, conditions, etc. 4
projects contain both 16S and mWGS components NCBI BioProject 46305 |
Framework of Clinical Metadata
In addition to the generation of mWGS and/or 16S metagenomic sequence data, metadata about the human subjects was also collected. To protect subject privacy, those data are available to qualified researchers only through NCBI's dbGaP portal. Qualified researchers are defined as PI-level investigators at legitimate institutions who have described how they plan to use the data and can follow a series of precautions to safeguard patient privacy. Detailed information on accessing private data is available at NCBI dbGaP.
The following clinical metadata fields are the only fields available outside of dbGaP. These fields have been directly embedded in the sequence file metadata and are also available through the HMP Metadata Catalog:
- Unique subject ID
- Body site
- Sex (male/female)
- Visit number
No approval is necessary to access these metadata fields.
Accessing Data
All sequence data is openly available for download. To protect subject privacy, data has been filtered prior to release by NCBI using BMTagger to remove contaminating human sequence. Filtered human sequence is available to qualified researchers only through NCBI's dbGaP portal. See more about applying for qualified researcher status above.
There are two primary portals for accessing HMP1 genomic and metagenomic sequence data:
- The HMP Data Portal, developed and maintained by the HMP Data Analysis and Coordination Center (DACC), provides access to raw and value added datasets. The DACC also provides access to a set of published, pre-computed analysis sets through the legacy HMP1 Data Browser
- NCBI Human Microbiome Roadmap Project page (Accession: PRJNA43021) provides access to raw sequence data. Most of the raw sequence data generated by HMP1 resides at NCBI's Sequence Read Archive (SRA). From this link, one can navigate to project-level Bioproject pages containing links to all associated SRA experiments (SRX). Please note that Healthy cohort 16S sequences were submitted under two separate BioProjects, based on date of submission (pre vs post 2012 publication).
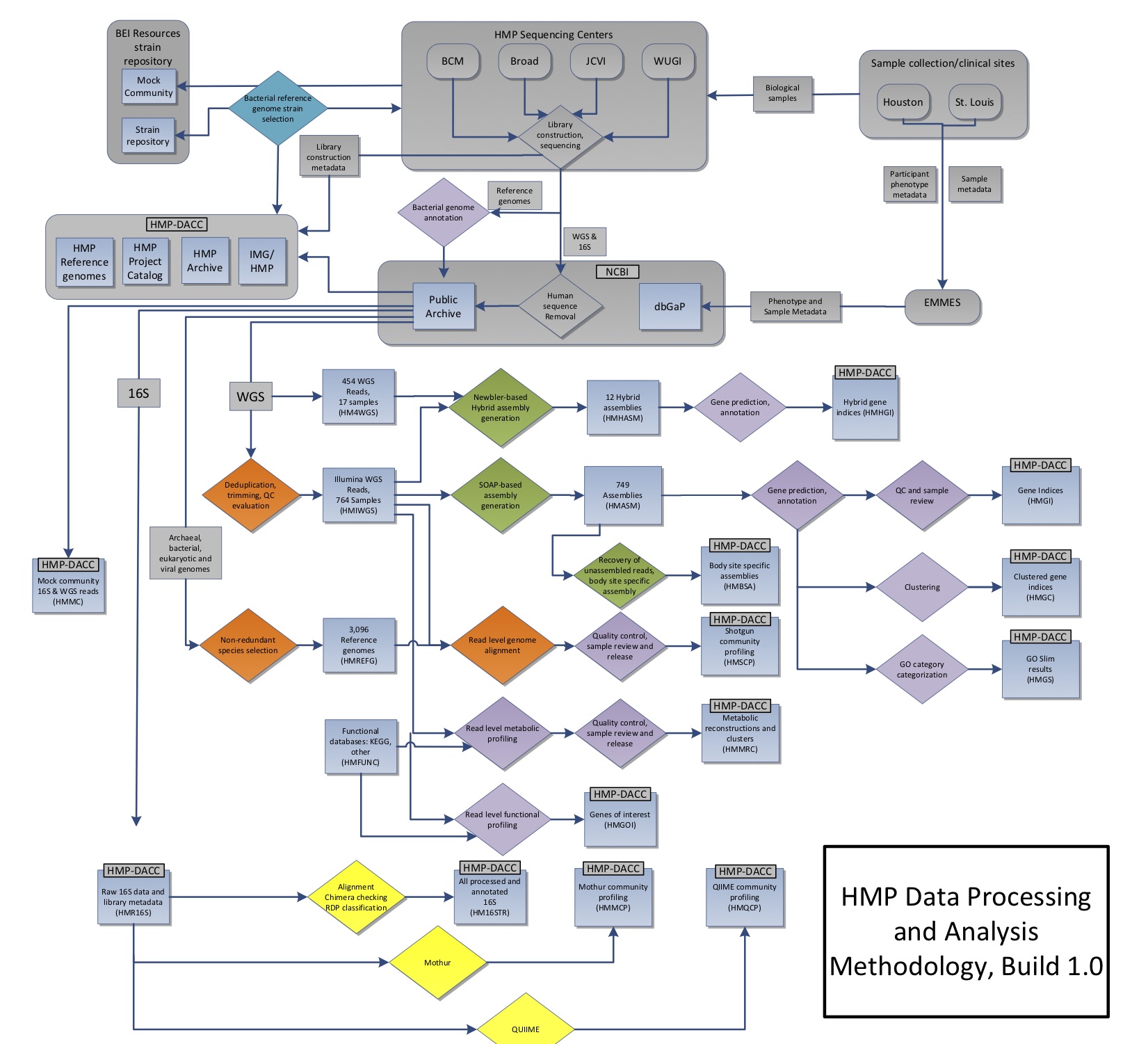
The diagram below shows the sampling and analysis flow for the initial set of analyzed HMP1 samples, as described and published in the Nature 2012 paper.